Cutoff selection¶
One of the most important tasks when constructing a cluster expansion is the choice of appropriate cutoffs. A simple way to select cutoffs is to scan a wide range of different values and monitor how the cross validation error changes, as will be demonstrated below.
Here, we will use the randomized dataset for the \(\ce{Mo_{1-x}V_xC_{1-y}}\) system.
[1]:
from ase.db import connect
from ase.io import read
# setup
chemical_symbols = [['C', 'Be'], ['Mo', 'V']]
db = connect('../dft-databases/rocksalt_MoVCvac_randomized.db')
prim = read('../structures/MoC_rocksalt_prim.xyz')
# collect training structures
training_structures = []
for row in db.select():
structure = row.toatoms()
structure.info['mixing_energy'] = row.mixing_energy
training_structures.append(structure)
[2]:
from icet import ClusterSpace, StructureContainer
def get_fit_data(cutoffs: list[float]):
""" Helper function for compiling the sensing matrix and
target mixing energies given a set of cutoffs. """
cs = ClusterSpace(prim, cutoffs, chemical_symbols)
sc = StructureContainer(cs)
for structure in training_structures:
sc.add_structure(
structure,
properties=dict(mixing_energy=structure.info['mixing_energy']))
return sc.get_fit_data(key='mixing_energy')
Selecting the pair cutoff¶
Next, we loop over different pairwise cutoffs and compute the cross-validation.
[3]:
import numpy as np
from pandas import DataFrame
from trainstation import CrossValidationEstimator
# parameters
fit_method = 'ardr'
train_size = 350
n_splits = 300
cutoff_pair_vals = [4, 5, 6, 7, 8, 9, 10, 11, 12]
# run pair cutoff scan
records = []
for c2 in cutoff_pair_vals:
print(f'pair cutoff: {c2:4.1f}')
A, y = get_fit_data([c2])
cve = CrossValidationEstimator(
(A, y), validation_method='shuffle-split', n_splits=n_splits,
fit_method=fit_method, train_size=train_size)
cve.train()
cve.validate()
records.append(dict(
cutoff_pair=c2,
rmse_train=cve.rmse_train,
rmse_validation=cve.rmse_validation,
n_parameters=cve.n_parameters,
))
df2 = DataFrame(records)
pair cutoff: 4.0
pair cutoff: 5.0
pair cutoff: 6.0
pair cutoff: 7.0
pair cutoff: 8.0
pair cutoff: 9.0
pair cutoff: 10.0
pair cutoff: 11.0
pair cutoff: 12.0
[4]:
from matplotlib import pyplot as plt
fig, ax = plt.subplots(
figsize=(4.5, 3.4),
dpi=120,
)
ax.plot(df2.cutoff_pair, 1e3 * df2.rmse_train,
'--s', label='training error')
ax.plot(df2.cutoff_pair, 1e3 * df2.rmse_validation,
'-o', label='validation error')
ax.set_xlabel('Pair cutoff (Å)')
ax.set_ylabel('RMSE (meV/site)')
ax.set_ylim(0, 15)
ax2 = ax.twinx()
ax2.plot(df2.cutoff_pair, df2.n_parameters,
':d', color='tab:red', label='number of parameters')
ax2.set_ylim(0, 60)
ax2.set_ylabel('Number of parameters')
ax.plot(np.nan, np.nan, ':d', color='tab:red', label='number of parameters')
ax.legend(loc='upper center')
fig.tight_layout()
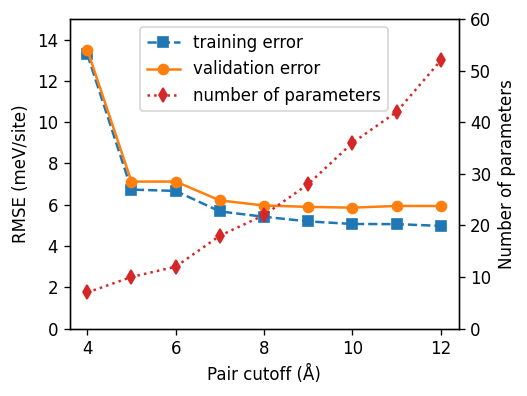
From the above figure it is apparent that the validation error does not improve beyond 9 Å. In fact it even increases slightly again. This indicates that a suitable choice for the pair cutoff is 9 Å.
Selecting the triplet cutoff¶
Next, we repeat the same procedure for the triplet cutoff using a fixed pair cutoff of 9 Å.
[5]:
cutoff_pair_final = 9.0
cutoff_triplet_vals = [3, 4, 5, 6, 7]
records = []
for c3 in cutoff_triplet_vals:
print(f'cutoff triplet: {c3}')
A, y = get_fit_data([cutoff_pair_final, c3])
cve = CrossValidationEstimator(
(A, y), validation_method='shuffle-split', n_splits=n_splits,
fit_method=fit_method, train_size=train_size)
cve.train()
cve.validate()
records.append(dict(
cutoff_triplet=c3,
rmse_train=cve.rmse_train,
rmse_validation=cve.rmse_validation,
n_parameters=cve.n_parameters,
))
df3 = DataFrame(records)
cutoff triplet: 3
cutoff triplet: 4
cutoff triplet: 5
cutoff triplet: 6
cutoff triplet: 7
[6]:
fig, ax = plt.subplots(
figsize=(4.5, 3.4),
dpi=120,
)
ax.plot(df3.cutoff_triplet, 1e3 * df3.rmse_train,
'--s', label='training error')
ax.plot(df3.cutoff_triplet, 1e3 * df3.rmse_validation,
'-o', label='validation error')
ax.set_ylim(0, 10)
ax.set_xlabel('Triplet cutoff (Å)')
ax.set_ylabel('RMSE (meV/site)')
ax2 = ax.twinx()
ax2.plot(df3.cutoff_triplet, df3.n_parameters,
':d', color='tab:red', label='number of parameters')
ax2.set_ylim(0, 150)
ax2.set_ylabel('Number of parameters')
ax.plot(np.nan, np.nan, ':d', color='tab:red', label='number of parameters')
ax.legend(loc='upper left')
fig.tight_layout()
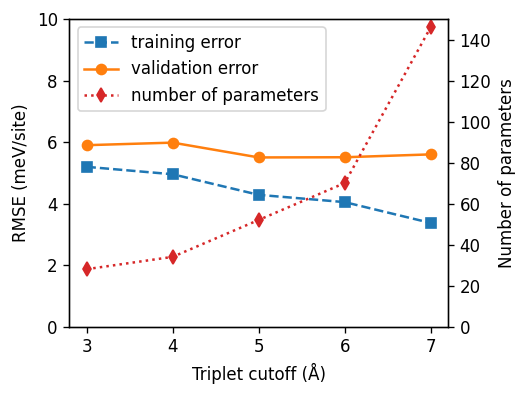
Applying the same logic as in the case of the pair cutoff this analysis indicates a triplet cutoff of 5 Å. This process can be repeated for higher order clusters (quadruplets etc) but in the present case this will not lead to a significant improvement of the model.
In general simpler models (shorter cutoffs) tend to yield more transferable and well-behaved models. In general, selecting cutoffs is not an exact procedure and one needs to balance accuracy vs simplicity of the model.