Additional material¶
Convergence of simulations¶
[784]:
from mchammer.data_analysis import analyze_data
fig, axes = plt.subplots(
figsize=(5.8, 4.6),
dpi=120,
nrows=3,
ncols=3,
sharex=True,
sharey='row',
)
temperature = 300
dmu, phi = 0.140, -1.250
dmu, phi = 0.120, -1.175
#dmu, phi = 0.000, -0.775
kappa = 50
# Number of MC cyclces to remove from the beginning of
# the trajectory to account for equilibration.
mcc_cutoff = 10
for icol, size in enumerate([4, 6, 8]):
for irow, ensemble in enumerate(['sgc', 'vcsgc']):
# load data container
if ensemble == 'sgc':
fname = f'runs/sgc-T{temperature}-dmu{dmu:.3f}-size{size}.dc'
else:
fname = f'runs/vcsgc-T{temperature}-phi{phi:.3f}-kappa{kappa}-size{size}.dc'
dc = DataContainer.read(fname)
# add columns with MC cycle and concentration
df = dc.data
n_sites = len(dc.structure)
df['mccycle'] = df.mctrial / n_sites
df['conc'] = df.Pd_count / n_sites
# remove equilibration period
df = df[df.mccycle > mcc_cutoff]
# compute averages for trajectories of different length (nmax)
data = []
for nmax in range(mcc_cutoff + 5, 100 + 1, 5):
record = dict(nmax=nmax)
data.append(record)
res = analyze_data(df[df.mccycle <= nmax].conc)
record.update({f'conc_{k}': v for k, v in res.items()})
if ensemble == 'vcsgc':
res = analyze_data(df[df.mccycle <= nmax].free_energy_derivative_Pd)
record.update({f'dfreen_{k}': v for k, v in res.items()})
df_avg = DataFrame.from_dict(data)
# plot mean concentration and error estimate
ax = axes[irow][icol]
color = 'C0' if ensemble == 'sgc' else 'C1'
ax.text(0.07, 0.85, f'{ensemble.upper()} {n_sites} atoms', transform=ax.transAxes)
ax.plot(df.mccycle, 1e2 * df.conc, color='0.5', alpha=0.3)
ax.plot(df_avg.nmax, 1e2 * df_avg.conc_mean, color=color)
ax.fill_between(df_avg.nmax,
y1=1e2 * (df_avg.conc_mean - df_avg.conc_error_estimate),
y2=1e2 * (df_avg.conc_mean + df_avg.conc_error_estimate),
color=color, alpha=0.2)
# plot mean free energy derivative and error estimate (VCGSC only)
if ensemble == 'vcsgc':
color = 'C1'
ax = axes[2][icol]
ax.plot(df_avg.nmax, df_avg.dfreen_mean, color=color)
ax.fill_between(df_avg.nmax,
y1=df_avg.dfreen_mean - df_avg.dfreen_error_estimate,
y2=df_avg.dfreen_mean + df_avg.dfreen_error_estimate,
color=color, alpha=0.2)
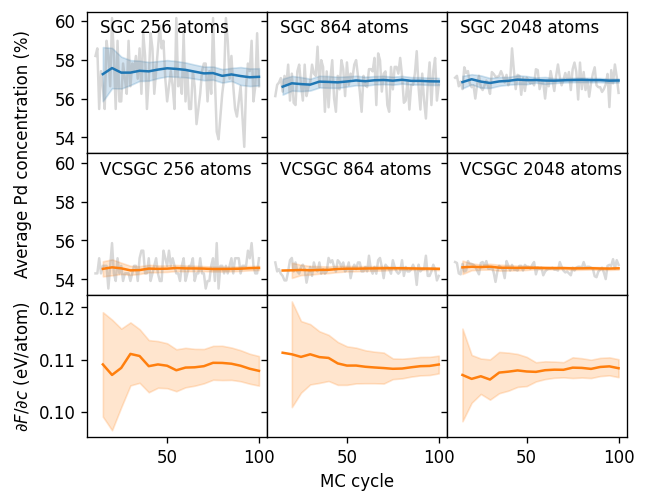
axes[1][0].set_ylim(axes[0][0].get_ylim())
axes[1][0].set_ylabel(r'Average Pd concentration (%)', y=1)
axes[2][0].set_ylabel(r'$\partial F/\partial c$ (eV/atom)')
axes[-1][1].set_xlabel(r'MC cycle')
fig.subplots_adjust(wspace=0, hspace=0)
fig.align_ylabels()
The VCSCG variance constraint \(\bar{\kappa}\)¶
The VCSGC ensemble requires one to specify both an average and a variance constraint for the concentration. So far we have used the recommended default value of \(\bar{\kappa}=200\). It is now instructive to explore this aspect in a bit more detail. To this end, we first repeat the previous VCSGC-MC simulations for several different values of \(\bar{\kappa}\).
[15]:
%%time
for kappa in [5, 10, 50, 500]:
for phi in phis:
mc = VCSGCEnsemble(
supercell,
calculator=calculator,
temperature=temperature,
phis={'Pd': phi},
kappa=kappa,
)
mc.run(len(supercell) * mc_cycles)
dcs_vcsgc[(phi, kappa)] = mc.data_container
CPU times: user 1min 22s, sys: 264 ms, total: 1min 22s
Wall time: 1min 23s
We compile the results in a DataFrame for convenient analysis.
[16]:
data = []
for (phi, kappa), dc in dcs_vcsgc.items():
n_sites = len(dc.structure)
Pd_count = dc.analyze_data('Pd_count', start=mcc_cutoff * n_sites)
accratio = dc.analyze_data('acceptance_ratio', start=mcc_cutoff * n_sites)
dfreen = dc.analyze_data(
'free_energy_derivative_Pd', start=mcc_cutoff * n_sites)
data.append(dict(
phi=phi,
kappa=kappa,
Pd_conc=Pd_count['mean'] / n_sites,
Pd_conc_err=Pd_count['error_estimate'] / n_sites,
Pd_conc_std=Pd_count['std'] / n_sites,
Pd_conc_corr=Pd_count['correlation_length'],
acceptance_ratio=accratio['mean'],
free_energy_derivative=dfreen['mean'],
free_energy_derivative_err=dfreen['error_estimate'],
))
df_vcsgc = DataFrame.from_dict(data)
[741]:
%%time
from glob import glob
files = sorted(glob('runs/vcsgc-T300-phi*-kappa2-size6.dc'))
df_vcsgc = collect_data(files)
100%|██████████| 248/248 [00:18<00:00, 13.09it/s]
CPU times: user 18.7 s, sys: 80.6 ms, total: 18.8 s
Wall time: 19 s
Below we plot the results, which allows us to make several relevant observations.
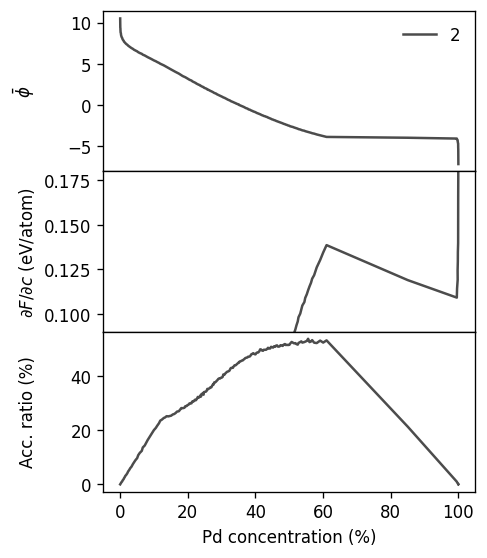
In VCSGC-MC simulations one obtains a nearly linear mapping between the average constraint parameter \(\bar{\phi}\) and the average concentration \(\left<c\right>\). This is consistent with the observation made above that the VCSGC-MC simulations yielded a more even sampling of the concentration range than the SGC-MC simulations.
When increasing \(\bar{\kappa}\), the concentration range sampled for a fixed range of \(\bar{\phi}\) increases. When \(\bar{\kappa}\) approaches zero on the other hand, the VCSGC ensemble maps back to the SGC ensemble.
The free energy derivative is independent of the choice of \(\bar{\kappa}\).
The acceptance ratio drops, however, with increasing \(\bar{\kappa}\), which means that longer runs are needed to achieve convergence.
[755]:
fig, axes = plt.subplots(
figsize=(4, 5.2),
dpi=120,
nrows=3,
sharex=True,
)
log_kappa_min = np.log(df_vcsgc.kappa.min())
log_kappa_max = np.log(df_vcsgc.kappa.max())
for kappa, df in df_vcsgc.groupby('kappa'):
if kappa < 5:
continue
kwargs = dict(
alpha=0.7,
label=f'{kappa}' if kappa in [2, 50, 500] else '',
color=cmap((np.log(kappa) - log_kappa_min) / (log_kappa_max - log_kappa_min)),
)
axes[0].plot(1e2 * df.conc_mean, df.phi, **kwargs)
axes[1].plot(1e2 * df.conc_mean, df.free_energy_derivative_Pd_mean, **kwargs)
axes[2].plot(1e2 * df.conc_mean, 1e2 * df.acceptance_ratio_mean, **kwargs)
axes[-1].set_xlabel(r'Pd concentration (%)')
axes[0].set_ylabel(r'$\bar{\phi}$')
axes[1].set_ylabel(r'$\partial F/\partial c$ (eV/atom)')
axes[2].set_ylabel(r'Acc. ratio (%)')
axes[0].legend(ncol=3, frameon=False)
axes[1].set_ylim(0.09, 0.18)
fig.align_ylabels()
fig.subplots_adjust(hspace=0)
[722]:
fig, axes = plt.subplots(
figsize=(4, 5.2),
dpi=120,
nrows=3,
sharex=True,
)
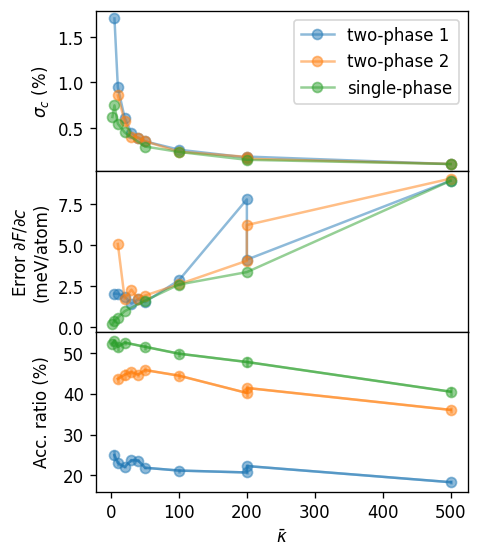
for k, (label, target_conc, tol) in enumerate([
('two-phase 1', 0.865, 0.01),
('two-phase 2', 0.726, 0.015),
('single-phase', 0.597, 0.004),
]):
df = df_vcsgc[np.isclose(df_vcsgc.conc_mean, target_conc, atol=tol)].sort_values('kappa')
df = df[df.kappa <= 500]
kwargs = dict(
color=f'C{k}',
alpha=0.5,
label=label,
)
ax = axes[0]
ax.plot(df.kappa, 1e2 * df.conc_std, 'o-', **kwargs)
ax.set_ylabel(r'$\sigma_c$ (%)')
ax = axes[1]
ax.plot(df.kappa, 1e3 * df.free_energy_derivative_Pd_error_estimate, 'o-', **kwargs)
ax.set_ylabel(r'Error $\partial F/\partial c$' + '\n(meV/atom)')
ax = axes[2]
"""
if '2' in label:
ax.plot(df.kappa, 1e2 * df.conc_mean, 'o-', **kwargs)
"""
ax.plot(df.kappa, 1e2 * df.acceptance_ratio_mean, 'o-', **kwargs)
ax.errorbar(df.kappa, 1e2 * df.acceptance_ratio_mean,
yerr=1e2 * df.acceptance_ratio_error_estimate, **kwargs)
ax.set_ylabel(r'Acc. ratio (%)')
axes[0].legend()
axes[-1].set_xlabel(r'$\bar{\kappa}$')
fig.align_ylabels()
fig.subplots_adjust(hspace=0)
[723]:
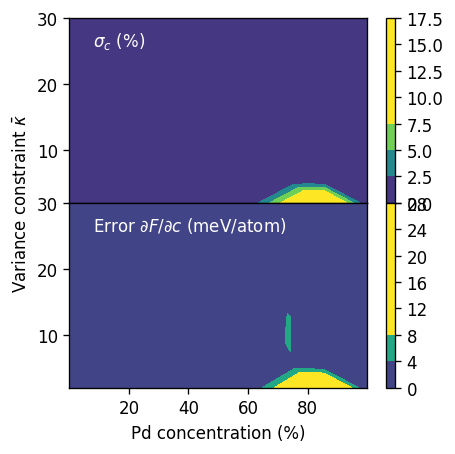
fig, axes = plt.subplots(
figsize=(4, 4),
dpi=120,
nrows=2,
sharex=True,
sharey=True,
)
df = df_vcsgc.dropna(subset='free_energy_derivative_Pd_error_estimate')
df = df[df.kappa <= 30]
#df = df[(df.conc_mean >= 0.6) & (df.conc_mean <= 0.9)]
ax = axes[0]
cm = ax.tricontourf(1e2 * df.conc_mean, df.kappa, 1e2 * df.conc_std, vmax=8)
plt.colorbar(cm, ax=ax)
pos = (0.08, 0.85)
ax.text(*pos, r'$\sigma_c$ (%)', transform=ax.transAxes, color='white')
ax = axes[1]
cm = ax.tricontourf(1e2 * df.conc_mean, df.kappa,
1e3 * df.free_energy_derivative_Pd_error_estimate, vmax=10)
plt.colorbar(cm, ax=ax)
ax.text(*pos, r'Error $\partial F/\partial c$ (meV/atom)', transform=ax.transAxes, color='white')
axes[-1].set_xlabel(r'Pd concentration (%)')
axes[1].set_ylabel(r'Variance constraint $\bar{\kappa}$', y=1)
fig.align_ylabels()
fig.subplots_adjust(hspace=0)
[ ]:
[ ]:
[ ]:
[ ]:
[ ]:
[406]:
from scipy.optimize import curve_fit
def rk_func(x, *pars):
xmod = (x - pars[0]) / (1 - pars[0])
f = 0
for k, p in enumerate(pars[1:]):
f += p * (1 - 2 * xmod) ** k * xmod * (1 - xmod)
return f
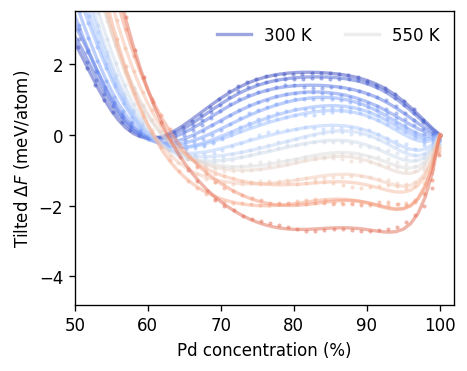
fig, ax = plt.subplots(
figsize=(4, 3.2),
dpi=120,
)
rk_order = 6
parameters = {}
for temperature, df in df_selection.groupby('temperature'):
try:
slope = boundaries_direct[boundaries_direct.temperature == temperature].iloc[0].slope
except:
continue
df.dropna(subset='freen', inplace=True)
df['freen_tilted'] = df.freen + slope * (1 - df.conc_mean)
df = df[df.conc_mean > 0.5]
pars, _ = curve_fit(rk_func, df.conc_mean, df.freen_tilted, p0=[0.6] + rk_order * [1])
parameters[temperature] = pars
color = cmap((temperature - tmin) / (tmax - tmin))
ax.scatter(1e2 * df.conc_mean, df.freen_tilted, alpha=0.5, s=6, lw=0, color=color)
xs = np.linspace(0, 1, 101)
ax.plot(1e2 * xs, rk_func(xs, *pars),
label=f'{temperature:.0f} K' if temperature in [tmin, 550, 750, tmax] else '',
alpha=0.5, lw=2, color=color)
ax.set_xlabel(r'Pd concentration (%)')
ax.set_ylabel(r'Tilted $\Delta F$ (meV/atom)')
ax.set_xlim(50, 102)
ax.set_ylim(-4.8, 3.5)
ax.legend(frameon=False, ncol=3)
fig.tight_layout()
[407]:
from icet.tools import ConvexHull
xs = np.linspace(0.5, 1, 101)
data = []
for temperature, pars in parameters.items():
ys = rk_func(xs, *pars)
ch = ConvexHull(xs, ys)
df_hull = DataFrame(np.array([ch.concentrations, ch.energies]).T,
columns=['conc_mean', 'freen'])
dfs = split_at_miscibility_gap(df_hull, clim=0.04)
if len(dfs) < 2:
continue
freen_left = dfs[-2].iloc[-1].freen
freen_right = dfs[-1].iloc[0].freen
conc_left = dfs[-2].iloc[-1].conc_mean
conc_right = dfs[-1].iloc[0].conc_mean
slope = (freen_right - freen_left) / (conc_right - conc_left)
data.append(dict(
temperature=temperature,
conc_left=conc_left,
conc_right=conc_right,
slope=slope,
))
boundaries_interpolated = DataFrame.from_dict(data)
[410]:
fig, ax = plt.subplots(
figsize=(4, 3.2),
dpi=120,
)
kwargs = dict(alpha=0.5, marker='o', color='C0')
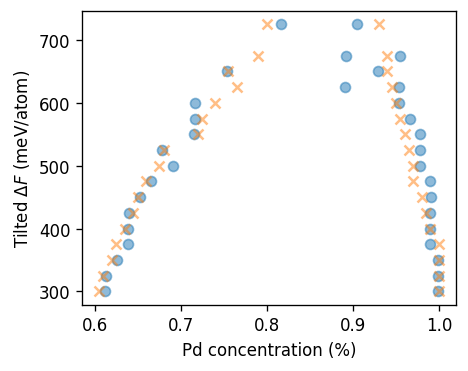
ax.scatter(boundaries_direct.conc_left,
boundaries_direct.temperature, **kwargs)
ax.scatter(boundaries_direct.conc_right,
boundaries_direct.temperature, **kwargs)
kwargs = dict(alpha=0.5, marker='x', color='C1')
ax.scatter(boundaries_interpolated.conc_left,
boundaries_interpolated.temperature, **kwargs)
ax.scatter(boundaries_interpolated.conc_right,
boundaries_interpolated.temperature, **kwargs)
ax.set_xlabel(r'Pd concentration (%)')
ax.set_ylabel(r'Tilted $\Delta F$ (meV/atom)')
#ax.set_xlim(50, 102)
#ax.set_ylim(-4.8, 3.5)
ax.legend(frameon=False, ncol=3)
fig.tight_layout()
No artists with labels found to put in legend. Note that artists whose label start with an underscore are ignored when legend() is called with no argument.